First step in Genome wide Analysis
Understanding Sequence Retrieval
The first step in performing genome wide analysis is the retrieval of sequences. This crucial process lays the foundation for further research and analysis. Without accurate sequence data, the results of any bioinformatics study may be flawed or misleading.
Selecting Your Crop and Domain
Begin by selecting the crop whose genome has been completely sequenced. This is essential as it will guide you in choosing the right data for your analysis. Next, identify the peptide sequence of the domain relevant to your research. Whether you aim to explore protein functions or gene regulation, pinpointing the appropriate domain is fundamental.
Retrieving the Sequences
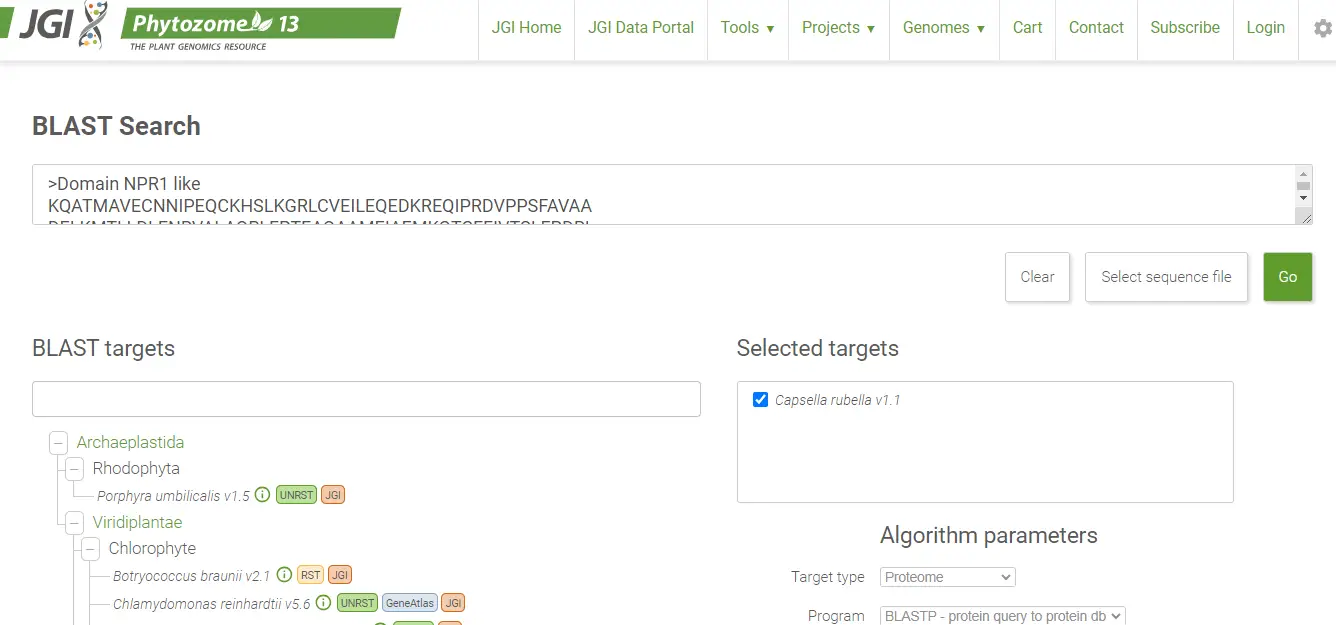
Next, visit genomic data repositories such as Phytozome or Plant Ensembl, among others, to locate the genome data for your selected crop. Afterward, access the BLAST tool on these platforms. Input your domain sequence and select the option to perform a protein BLAST search. Upon completion, download the protein or peptide sequences and save the file as ‘raw’ for future reference.
It’s imperative to rename the file for easier identification. Repeat these steps for any additional sequences you need, including genomic data if applicable. By following these steps meticulously, you will ensure that you have the necessary sequences to proceed with your genome wide analysis effectively.
Steps for the retrieval of sequences for Genome wide analysis
Following sub-steps are followed in this step.
1. First select the crop whose genome is sequenced completely.
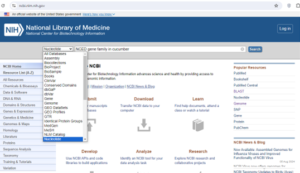
 https://phytozome-next.jgi.doe.gov/info/Ahypochondriacus_v2_1
https://phytozome-next.jgi.doe.gov/info/Ahypochondriacus_v2_1
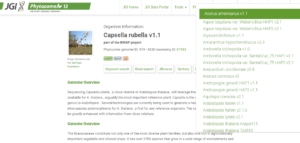
2. Now you need to select the peptide sequence of the domain you are working, or you want to work.
3. Go to websites like (Phytozome, Plant Ensemble, or any other website containing genome data of your crop.
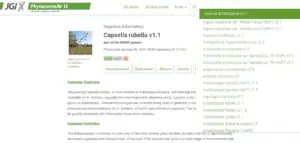
https://phytozome-next.jgi.doe.gov/info/Crubella_v1_1
4. Now select the crop on the website and go to blast.
5. Add your domain sequence select the option blast proteins and blast the sequence.
>NP_176610.1 regulatory protein (NPR1) [Arabidopsis thaliana]
MDTTIDGFADSYEISSTSFVATDNTDSSIVYLAAEQVLTGPDVSALQLLSNSFESVFDSPDDFYSDAKLV
LSDGREVSFHRCVLSARSSFFKSALAAAKKEKDSNNTAAVKLELKEIAKDYEVGFDSVVTVLAYVYSSRV
RPPPKGVSECADENCCHVACRPAVDFMLEVLYLAFIFKIPELITLYQRHLLDVVDKVVIEDTLVILKLAN
ICGKACMKLLDRCKEIIVKSNVDMVSLEKSLPEELVKEIIDRRKELGLEVPKVKKHVSNVHKALDSDDIE
LVKLLLKEDHTNLDDACALHFAVAYCNVKTATDLLKLDLADVNHRNPRGYTVLHVAAMRKEPQLILSLLE
KGASASEATLEGRTALMIAKQATMAVECNNIPEQCKHSLKGRLCVEILEQEDKREQIPRDVPPSFAVAAD
ELKMTLLDLENRVALAQRLFPTEAQAAMEIAEMKGTCEFIVTSLEPDRLTGTKRTSPGVKIAPFRILEEH
QSRLKALSKTVELGKRFFPRCSAVLDQIMNCEDLTQLACGEDDTAEKRLQKKQRYMEIQETLKKAFSEDN
LELGNSSLTDSTSSTSKSTGGKRSNRKLSHRRR
>Domain NPR1 like
KQATMAVECNNIPEQCKHSLKGRLCVEILEQEDKREQIPRDVPPSFAVAA
DELKMTLLDLENRVALAQRLFPTEAQAAMEIAEMKGTCEFIVTSLEPDRL
TGTKRTSPGVKIAPFRILEEHQSRLKALSKTVELGKRFFPRCSAVLDQIM
NCEDLTQLACGEDDTAEKRLQKKQRYMEIQETLKKAFSEDNLELGNSSLT
DSTSST

https://phytozome-next.jgi.doe.gov/blast-search
6. Now download your protein/peptide sequences and save the file as RAW.
https://phytozome-next.jgi.doe.gov/blast-results/1230931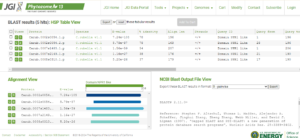
7. Rename the file.

8. Same steps are followed to download remaining sequences. i.e., Genomic & CDS sequences of each gene.
9. For the Promoter sequences select the 1000 up sequence of the respective genes.



Motif And Domain Analysis Of Protein In Genome Ide Analysis
[…] The First Step in Genome Wide Analysis: Sequence Retrieval […]
Genome Wide Analysis Tools: TBTool, Mega X, And EndNote
[…] The First Step in Genome Wide Analysis: Sequence Retrieval […]
A Step-by-Step Guide For Genome-Wide Analysis Of Gene Families In Plants
[…] The First Step in Genome Wide Analysis: Sequence Retrieval […]