Introduction to RNAi and C. elegans
RNA interference (RNAi) is biological process that regulates gene expression and provides a defense mechanism against viral infections and transposable elements in various organisms including plants. This post-transcriptional gene silencing (PTGS) process involves the degradation of messenger RNA (mRNA) molecules, which effectively prevents the translation of specific genes into proteins. The key players in RNAi are double-stranded RNA (dsRNA) molecules, which can be derived from natural sources or artificially synthesized. Understanding RNAi at a molecular level reveals its potential applications in research and biotechnology.
At the core of RNAi is the processing of dsRNA into smaller fragments known as small interfering RNAs (siRNAs). These siRNAs are then incorporated into the RNA-induced silencing complex (RISC), where they guide the complex to complementary mRNA strands. The binding of siRNA to mRNA leads to the cleavage of the mRNA, thereby silencing the corresponding gene. This mechanism relies on both the sense and antisense strands of dsRNA. The sense strand typically matches the mRNA sequence, while the antisense strand is complementary, allowing for precise targeting and regulation at the genetic level.
Caenorhabditis elegans, commonly known as C. elegans, serves as a premier model organism for studying RNAi. This nematode exhibits well-characterized genetics, a short life cycle, and a simple anatomy, making it advantageous for research. Notably, C. elegans can uptake dsRNA through its intestinal epithelium, allowing for efficient RNAi studies. Research has shown that the RNAi pathway in C. elegans shares similarities with higher organisms, reinforcing its significance in the broader field of genetics and molecular biology. Findings from C. elegans studies continue to influence our understanding of gene regulation and hold promise for advancements in agricultural biotechnology and therapeutic interventions.
Historical Perspectives: Jorgensen to Baulcombe
Jorgensen’s findings laid the foundation for understanding gene silencing mechanisms. Following his work, David Baulcombe made notable strides in exploring RNAi in plants. Baulcombe’s research highlighted the critical role of small interfering RNAs (siRNAs) in the process of gene silencing, which further enriched the body of knowledge surrounding RNAi. His approach demonstrated that the introduction of dsRNA could lead to the degradation of complementary mRNA in plant cells, thereby inhibiting the expression of targeted genes. This helped clarify the biochemical pathways involved in RNA interference and established a clear link between dsRNA and practical gene regulation in diverse organisms.
The transition from fundamental research to applied science was evident as the implications of RNAi began to flourish. The understanding that RNAi could be harnessed for agricultural purposes, such as developing pest-resistant crops or enhancing plant stress tolerance, significant advances in horticulture were made. Over the years, the exploration of RNAi mechanisms has evolved and expanded to encompass various organisms, revealing a universal biological strategy for regulating gene expression. Such developments underscore how the early experiments by Jorgensen and Baulcombe have transformed the landscape of molecular biology and agricultural science.
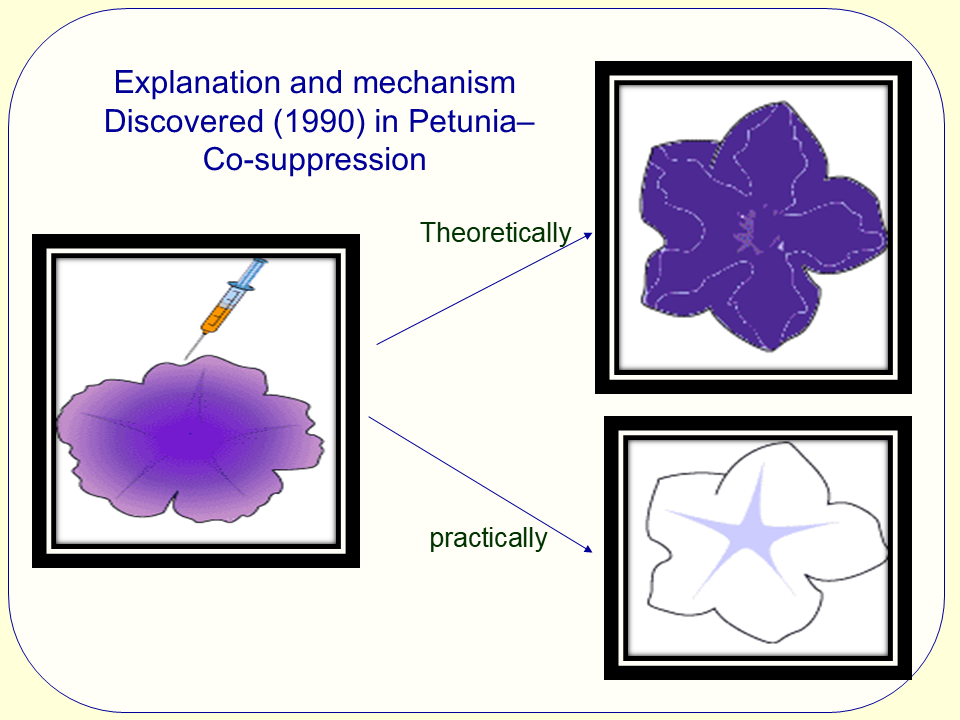
The Science of Co-suppression in C. elegans
Co-suppression, a remarkable phenomenon observed in Caenorhabditis elegans, plays a crucial role in gene regulation and has significant implications for the study of RNA interference (RNAi). This occurrence arises when the expression of a gene is inhibited not only in the presence of a specific transgene but also in other homologous genes within the organism. At a genetic level, co-suppression is mediated by the introduction of double-stranded RNA (dsRNA), which triggers a complex cascade of events involving the RNA-induced silencing complex (RISC). The resulting cleavage of target mRNA leads to the downregulation of both the target gene and its related homologs.
The primary mechanism underlying co-suppression involves the pathway of RNAi, where dsRNA derived from the transgene activates Dicer, an endonuclease responsible for cutting the dsRNA into small interfering RNAs (siRNAs). These siRNAs guide RISC to complementary mRNA molecules, promoting their degradation. Interestingly, when the dsRNA shares sequence homology with other genes, the silencing effect spreads, leading to the unintended suppression of alleles that share similar sequences. This feature of co-suppression not only highlights the evolutionary advantages of RNAi pathways but also introduces complexities in experimental design when conducting RNAi studies.
Several experimental examples illustrate the application and consequences of co-suppression in C. elegans. For instance, researchers have demonstrated the co-suppression of a neuropeptide gene by introducing a dsRNA construct related to another neuropeptide, resulting in significant alterations in behavioral phenotypes. These findings underscore the necessity for careful consideration in the interpretation of results from RNAi experiments, as co-suppression can often confound the effects attributed to targeted gene silencing. Thus, understanding co-suppression is essential for enhancing the accuracy and reliability of gene function studies in this model organism.
Horticultural Applications of siRNA and RNAi Technology
Small interfering RNA (siRNA) and RNA interference (RNAi) technology have emerged as pivotal tools in horticulture, providing innovative solutions for enhancing crop resilience and quality. These applications leverage the natural mechanisms of gene regulation to target specific pests, diseases, and undesirable plant traits, thereby advancing agricultural practices significantly. One of the primary horticultural benefits of RNAi is its ability to confer resistance against various pathogens. For instance, siRNA can effectively target and silence the genes responsible for susceptibility to viruses and fungal infections, resulting in healthier crop yields.
Another critical application of RNAi technology in horticulture is the improvement of plant traits, such as fruit quality, shelf life, and stress tolerance. Researchers are using this technology to enhance nutritional profiles, increase resistance to abiotic stressors like drought, and augment the flavors and textures of fruits and vegetables. For example, specific projects have targeted the pathways involved in sugar and acid accumulation in tomatoes, resulting in sweeter and more flavorful produce, a crucial factor for market competitiveness.
Despite its potential, the implementation of RNAi technology in horticulture faces various challenges. Regulatory frameworks are still in the developmental stage across many regions, which can hamper the swift adoption of these innovations. Additionally, the environmental impact and the long-term efficacy of genetically modified plants using RNAi remain under scrutiny. Current research trends are addressing these concerns, focusing on precision agriculture practices, which aim to minimize off-target effects while maximizing the benefits of RNAi applications.
Looking ahead, the future of siRNA and RNAi technology in horticulture appears promising. With continued advances in gene editing tools and deeper understanding of plant genomes, there is potential for developing even more robust cultivation strategies. As ongoing projects demonstrate the tangible effects of RNAi on specific plants, its role in sustainable agriculture may continue to expand, marking a transformative period in the field of horticulture.
Power Point Presentations






The Industrial Application Of RNAi In Horticultural Crops
[…] Exploring RNA Interference and Horticultural Applications […]