Introduction to Southern Hybridization
Southern Hybridization (SH), is a important technique in the field of molecular biology. Named after SH inventor, the British biologist Edwin Southern, this method was first developed in the 1970s. The pioneering work of SH revolutionized the way scientists detect specific sequences of DNA within complex mixtures, laying the ground work for many advancements in genetic research.
 Southern’ hybridization named after Sir Edwin Southern
Southern’ hybridization named after Sir Edwin Southern
The primary purpose of Southern Hybridization
- Immobilize DNA onto a permanent substrate
- Identify DNA sequence (gene) of interest
The identification and characterization of DNA fragments containing particular sequences. This is achieved by transferring DNA fragments from an agarose gel onto a membrane, followed by hybridization with a labeled DNA probe complementary to the target sequence. The selective nature of the probe ensures high specificity in detecting the desired DNA segments amidst a plethora of genomic content.
In the context of molecular plant science research, Southern Hybridization holds immense significance. Plants, with their vast and complex genomes, present unique challenges for genetic studies. Utilizing Southern blotting, researchers can effectively locate and analyze genes of interest, facilitating studies in gene expression, genetic mapping, and the investigation of gene function. This technique has been integral in advancing our understanding of plant genomics, contributing to innovations in crop improvement, disease resistance, and biotechnology applications.
The methodology has also been a cornerstone in various genetic applications, including the identification of genotypic variations, detection of transgenic plants, and examining evolutionarily conserved sequences across different species. The rigor and precision of Southern Hybridization have made it an indispensable tool in molecular plant science laboratories worldwide, ensuring accurate and reliable results that drive forward the field of genetic research. As molecular biology continues to evolve, the foundational techniques established by scientists like Edwin Southern remain crucial, setting the stage for future discoveries and advancements.
Principles Behind Southern Hybridization
Southern Hybridization is a pivotal molecular biology technique named after its inventor, Edwin Southern. This method hinges on several core scientific principles that collectively enable the detection of specific DNA fragments. Central to Southern Hybridization is the utilization of DNA restriction enzymes. These enzymes act like molecular scissors, cleaving DNA molecules at recognized sequences. This precise cutting produces distinct DNA fragments that can be analyzed further.
The next step in Southern Hybridization involves gel electrophoresis. This technique allows the separation of DNA fragments based on size and charge. By applying an electric field, DNA fragments migrate through an agarose gel matrix, with smaller fragments moving faster than larger ones. This separation forms a distinguishable pattern of bands, which can be visualized under UV light when stained with a DNA-binding dye.
Following electrophoresis, the separated DNA fragments are transferred—or blotted—onto a membrane, typically made of nylon or nitrocellulose, in a process aptly named blotting. The transfer can be carried out through capillary action or electrophoretic transfer, which ensures that the DNA fragments maintain their unique positions from the gel. This membrane serves as a stable platform for subsequent hybridization, offering enhanced sensitivity and durability compared to the gel.
Hybridization is a crucial step wherein labeled probes, often DNA or RNA sequences complementary to the target DNA fragments, are introduced to the membrane. These probes are tagged with radioactive or fluorescent labels, facilitating their detection. The hybridization process capitalizes on the principle of complementary base pairing, enabling the probes to bind specifically to their matching DNA sequences on the membrane.
Together, these techniques—restriction enzyme digestion, gel electrophoresis, blotting, and hybridization—create a robust workflow for detecting specific DNA sequences. The significance of Southern Hybridization in molecular plant science research lies in its ability to analyze genetic information, such as gene presence, gene mapping, and genetic diversity. This comprehensive approach provides researchers with essential insights into plant genomes, advancing the field of molecular biology.
Materials and Equipment Needed
Successful Southern Hybridization necessitates a meticulously chosen collection of materials and equipment, playing a critical role in ensuring reliable and reproducible results in molecular plant science research. The primary reagents include restriction enzymes, which are pivotal for digesting DNA samples into manageable fragments. Given the specificity of these enzymes, it’s essential to select ones that generate consistent and accurate cuts.
Equally important are buffers, which maintain optimal pH and ionic strength conditions during various stages of the procedure. TAE (Tris-acetate-EDTA) and TBE (Tris-borate-EDTA) buffers are commonly used in electrophoresis, while SSC (sodium saline citrate) buffer plays a significant role during the hybridization step. High-quality, freshly prepared buffers prevent experimental variabilities and ensure effective interactions between the DNA and the probes.
An agarose gel is another fundamental component, utilized for the separation of DNA fragments via electrophoresis. Typically, gels of 0.7% to 1.2% concentration are prepared depending on the size of the DNA fragments to be resolved. Membrane materials, such as nylon or nitrocellulose, are employed to transfer DNA from the agarose gels. Nylon membranes, known for their durability and high sensitivity, are often preferred for their superior binding capacity.
Once the DNA has been successfully transferred, probes are used to detect specific sequences. Probes can be radioactively labeled or non-radioactively labeled, with the latter encompassing digoxigenin or biotin labels, which are safer alternatives. The choice of detection system—whether autoradiography for radioactive probes or chemiluminescence for enzyme-conjugated probes—should match the labeling method to ensure optimal sensitivity and specificity.
Additional apparatus includes the electrophoresis unit, blotting apparatus, and hybridization ovens or chambers. Lastly, high-resolution imaging systems are fundamental for the detection and analysis of hybridized probes, offering a detailed visual representation of the resultant molecular map. Each piece of equipment must be maintained and calibrated, underscoring the significance of precision in molecular plant science research rooted in Southern Hybridization techniques.
Step-by-Step Procedure with Visual Demonstrations
Southern Hybridization, an essential technique in molecular plant science research, involves several meticulous steps. Ensuring precision at each stage is crucial for accurate results. Below, we provide a detailed protocol, accompanied by practical tips and visual aids to facilitate your understanding.
DNA Extraction and Purification
Start by isolating DNA from plant tissue using a suitable extraction method, such as CTAB extraction. Ensure the DNA is pure by measuring the A260/A280 ratio. An optimal ratio is generally between 1.8 and 2.0.
 Step 1. DNA isolation from Plants
Step 1. DNA isolation from Plants
Restriction Digestion
Select restriction enzymes that generate fragment sizes compatible with your analysis. Incubate the purified DNA with these enzymes in the appropriate buffer at the recommended temperature. This step is critical as it segments the DNA into smaller, manageable pieces.
 Step 2. Restriction Enzyme Digestion
Step 2. Restriction Enzyme Digestion
Agarose Gel Electrophoresis
Prepare a gel of the appropriate agarose concentration (usually 0.8-1.0%). Load the digested DNA samples along with a DNA ladder onto the gel. Run the gel at a suitable voltage until the dye front has migrated to an optimal distance. This separation process will enable you to visualize the distinct DNA fragments.
 Step 3. Agarose Gel Electrophoresis
Step 3. Agarose Gel Electrophoresis
Denaturation
The double stranded DNA molecules in the gel are denatured, into single strands. This is typically achieved by soaking and shaking the gel in a denaturing solution, such as sodium hydroxide (NaOH) or formamide. The denaturing solution disrupts the hydrogen bonds between the complementary DNA strands, causing them to separate.
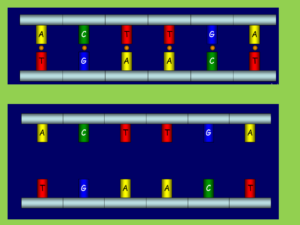 Step 4. Eliminate hydrogen bonds with sodium hydroxide (NaOH)
Step 4. Eliminate hydrogen bonds with sodium hydroxide (NaOH)
Blotting
Transfer the DNA fragments from the agarose gel onto a nylon or nitrocellulose membrane using a capillary or vacuum blotting system. Ensure thorough contact between the gel and the membrane to facilitate efficient transfer. Fix the DNA to the membrane via UV crosslinking or baking.
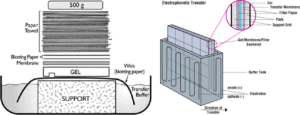 Step 5. Transfer DNA to Membrane
Step 5. Transfer DNA to Membrane
Hybridization
Prehybridize the membrane in hybridization buffer to block nonspecific binding sites. Then, incubate it with a labeled DNA probe complementary to the target sequence. The hybridization step ensures that specific DNA fragments are tagged for detection. Optimize hybridization conditions, such as temperature and ionic strength, to enhance specificity.
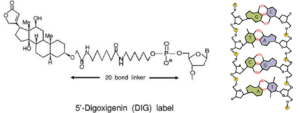 Step 6. Making a Probe
Step 6. Making a Probe
Detection
Following hybridization, wash the membrane to remove any unbound probe. The stringency of these washes is determined based on the probe’s properties and desired specificity. Detect the hybridized probe using an appropriate method, such as autoradiography or chemiluminescence, to visualize the target DNA fragments.
By adhering to each step meticulously and employing the outlined practical tips, you enhance the likelihood of successful Southern Hybridization. This technique, when executed correctly, can offer profound insights into plant genetic research, enabling the identification and characterization of specific DNA sequences.

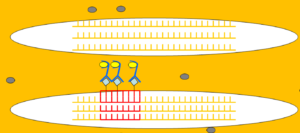 Step 7. Detection and Hybridization
Step 7. Detection and Hybridization
Data Analysis and Interpretation
Data analysis and interpretation are pivotal stages in Southern Hybridization experiments. The ultimate goal is to decipher the molecular information encoded within the hybridization patterns. Successful hybridization results typically manifest as distinct bands on an autoradiograph or chemiluminescent film. These bands correspond to the DNA fragments that have hybridized with the labeled probe.
Common patterns of hybridization include single or multiple bands, depending on the complexity of the genetic material under study. A single band is often indicative of a simple gene structure, whereas multiple bands may suggest the presence of gene families, repetitive sequences, or polymorphisms. Quantitative analysis requires densitometry, a technique that converts the intensity of the bands into numerical data, allowing for comparative analysis between different samples.
Interpretation of Southern Hybridization results necessitates careful consideration of various factors. Clear and strong signals usually signify successful hybridization, whereas weak or absent signals might indicate issues such as poor probe labeling, insufficient hybridization time, or degradation of the DNA. In instances of non-specific binding, where the probe hybridizes to non-target sequences, washing conditions must be optimized to improve specificity.
To ensure accurate interpretation, it is essential to include controls, such as positive controls with known hybridization outcomes and negative controls lacking the target sequence. These controls serve as benchmarks to assess the reliability of the experiment. Additionally, statistical analysis, like determining the signal-to-noise ratio, can enhance the confidence in the observed data.
For instance, in an experiment aiming to detect a single gene copy, a clear, sharp band at the expected position would confirm its presence. Conversely, the appearance of multiple bands might suggest gene duplication or the presence of restriction enzyme recognition site polymorphisms within the gene. In such cases, further analysis, like sequencing or using different probes, can provide more insights.
Ensuring accurate data interpretation involves a comprehensive understanding of molecular biology principles, meticulous experimental design, and prudent data analysis techniques. By integrating these elements, researchers can derive meaningful conclusions from Southern Hybridization experiments, significantly advancing molecular plant science research.
Applications of Southern Hybridization in Molecular Plant Science
Southern hybridization, a cornerstone of molecular plant science, has profoundly impacted various facets of plant research. This versatile technique offers unparalleled insights into plant genomics and has facilitated significant advancements in our understanding of plant biology.
One of the primary applications of Southern hybridization is in gene mapping. By utilizing this method, researchers can locate specific genes within a plant’s genome. This information is crucial for breeding programs and genetic improvement initiatives, allowing scientists to identify desirable traits and expedite the development of new plant varieties. Furthermore, gene mapping through Southern hybridization provides a foundational understanding of genetic linkages and sequence organization within plant genomes.
In addition to gene mapping, Southern hybridization is instrumental in the identification of transgenic plants. When plants are genetically modified, it’s essential to confirm the integration and stability of the introduced genes. Southern hybridization enables researchers to verify the presence of these genes and assess their copy number and integration sites within the plant genome. This reliability is critical for ensuring the success and safety of genetically modified crops.
Studying gene expression patterns is another significant application of Southern hybridization. By examining the presence and abundance of specific genomic sequences, researchers can infer patterns of gene expression across different tissues or developmental stages. This information is pivotal for understanding how genes regulate physiological processes and responses to environmental stimuli, ultimately advancing our knowledge of plant function and adaptation.
Moreover, Southern hybridization has proven invaluable in detecting plant pathogen infections. By identifying pathogen-specific DNA sequences, this technique allows for the early and accurate diagnosis of plant diseases. This capability is essential for effective disease management and the development of resistant plant varieties, thereby safeguarding crop yields and agricultural sustainability.
Through these diverse applications, Southern hybridization has become an indispensable tool in molecular plant science. Its contributions have not only advanced genetic research but also provided critical insights into the complexities of plant biology.
Comparison with Other Molecular Techniques
Southern Hybridization is one of several molecular techniques used in molecular plant science research. This method is specifically designed for the detection of specific DNA sequences within a DNA sample. The technique’s counterpart, Northern blotting, has similar principles but is used for RNA detection, allowing for the study of gene expression. While Northern blotting provides valuable insights into transcriptional activity, it does not capture genomic information, limiting its scope compared to Southern Hybridization.
Western blotting, on the other hand, is employed to detect specific proteins within a complex protein mixture. It relies on antibody-antigen interactions, making it a robust tool for studying protein expression and post-translational modifications. However, it does not provide information about the gene sequences encoding the proteins, thus offering a different angle of analysis compared to Southern Hybridization.
Polymerase Chain Reaction (PCR) is another widely used technique in molecular biology. It allows for the exponential amplification of specific DNA sequences, making it extremely sensitive and suitable for detecting minute amounts of DNA. PCR’s strength lies in its rapid amplification process and its ability to detect mutations or gene deletions accurately. However, PCR requires prior knowledge of the sequences to design primers, while Southern Hybridization can identify and isolate unknown DNA sequences based on hybridization patterns.
Southern Hybridization has unique aspects that make it indispensable in certain research scenarios. It excels in the detection of large-scale genomic rearrangements, gene insertions, deletions, and extensive structural variations that are often challenging to detect with PCR. This technique is particularly advantageous when studying gene families and repetitive DNA sequences where precise information about physical DNA arrangement is crucial.
In summary, while Northern blotting, Western blotting, and PCR each offer distinct advantages in molecular research, Southern Hybridization remains a critical tool for tasks requiring detailed DNA sequence analysis and genomic structure identification. Its ability to detect complex genetic variations makes it an essential technique for comprehensive molecular plant science research.
Future Directions and Innovations
As we continue to advance in molecular biology, the technique of Southern Hybridization is poised to undergo significant innovations, expanding its utility and improving its efficiency. Future improvements are likely to focus on enhancing sensitivity, speed, and reliability. One promising development is the integration of digital imaging systems and automated data analysis, which could drastically reduce the time required for results interpretation and allow for a higher throughput of samples.
Next-generation sequencing (NGS) technologies are also expected to play a critical role in evolving Southern Hybridization. These technologies provide in-depth insight into genetic material and offer a platform for developing more precise probes and hybridization protocols. Leveraging bioinformatics tools to design probes can enhance the specificity and sensitivity of Southern blot assays, enabling the detection of even low-abundance nucleic acids. Additionally, the use of fluorescent labels instead of traditional radioactive labels significantly improves safety and simplifies the detection process.
Nanotechnology is another area that shows potential for revolutionizing Southern Hybridization. Nanoparticles can be utilized to develop more efficient probing systems, which could lead to faster hybridization times and higher sensitivity. Moreover, advancements in microfluidics could reduce the amount of reagents and samples required, thus making the technique more cost-effective and practical for routine use in various research settings.
Ongoing research is actively exploring these avenues, aiming to refine the technique and broaden its applications. In plant science, for instance, novel applications could include more precise identification of gene expression patterns and more efficient marker-assisted selection in crop breeding programs. The technique’s utility could also extend to virology, microbiology, and environmental biology, where robust detection of specific DNA sequences plays a pivotal role.
The potential for innovation in Southern Hybridization is vast, promising to enhance our understanding of genetic information and drive progress across numerous fields of biological research. As new methodologies and technologies are adopted, Southern Hybridization will likely become an even more powerful tool in the molecular biologist’s toolkit.
*#SouthernBlot
*#MolecularBiology
*#DNAAnalysis
*#GeneticEngineering
*#Genomics
*#LabTechniques
*#MolecularTechniques
*#DNAHybridization
*#GeneticResearch
*#Biotechnology
*#molecularplantscience
*#CancerResearch
*#GeneticDisease
*#ForensicScience
*#PlantGenetics
*#Microbiology