Introduction to Gene Families in Plants
Gene families play a vital role in the genetic architecture of plants, encompassing groups of related genes that have arisen through the process of evolution from a common ancestor. These genes typically exhibit similar sequences and may carry out analogous functions, reflecting their shared evolutionary history. Understanding gene families is crucial for elucidating the underlying mechanisms of plant biology, as they are integral to various biological processes.

The significance of gene families extends beyond their evolutionary origin; they are fundamental to the adaptation and diversity observed in the plant kingdom. By analyzing gene families, researchers can gain insights into how plants have evolved in response to environmental pressures. For instance, certain gene families are involved in the development of specific traits that enhance survival and reproductive success, such as drought resistance or pest tolerance.

Moreover, gene families contribute to essential physiological and metabolic processes within plants. They are implicated in the synthesis of metabolites, which are crucial for growth, reproduction, and defense against biotic and abiotic stressors. The intricate relationships between gene families and these key processes highlight their relevance in research focusing on plant improvement and sustainability. For example, manipulating specific gene families may lead to enhanced stress tolerance in crops, thus improving yield and food security in the face of climate change.

In this context, exploring the diversity and evolution of gene families becomes essential to our understanding of the complex interactions that govern plant life. As genomic technologies advance, the study of gene families is positioned to provide deeper insights into the genetic basis of plant traits, contributing to significant agricultural advances and biodiversity conservation efforts.
Methodologies for Genome-Wide Analyses
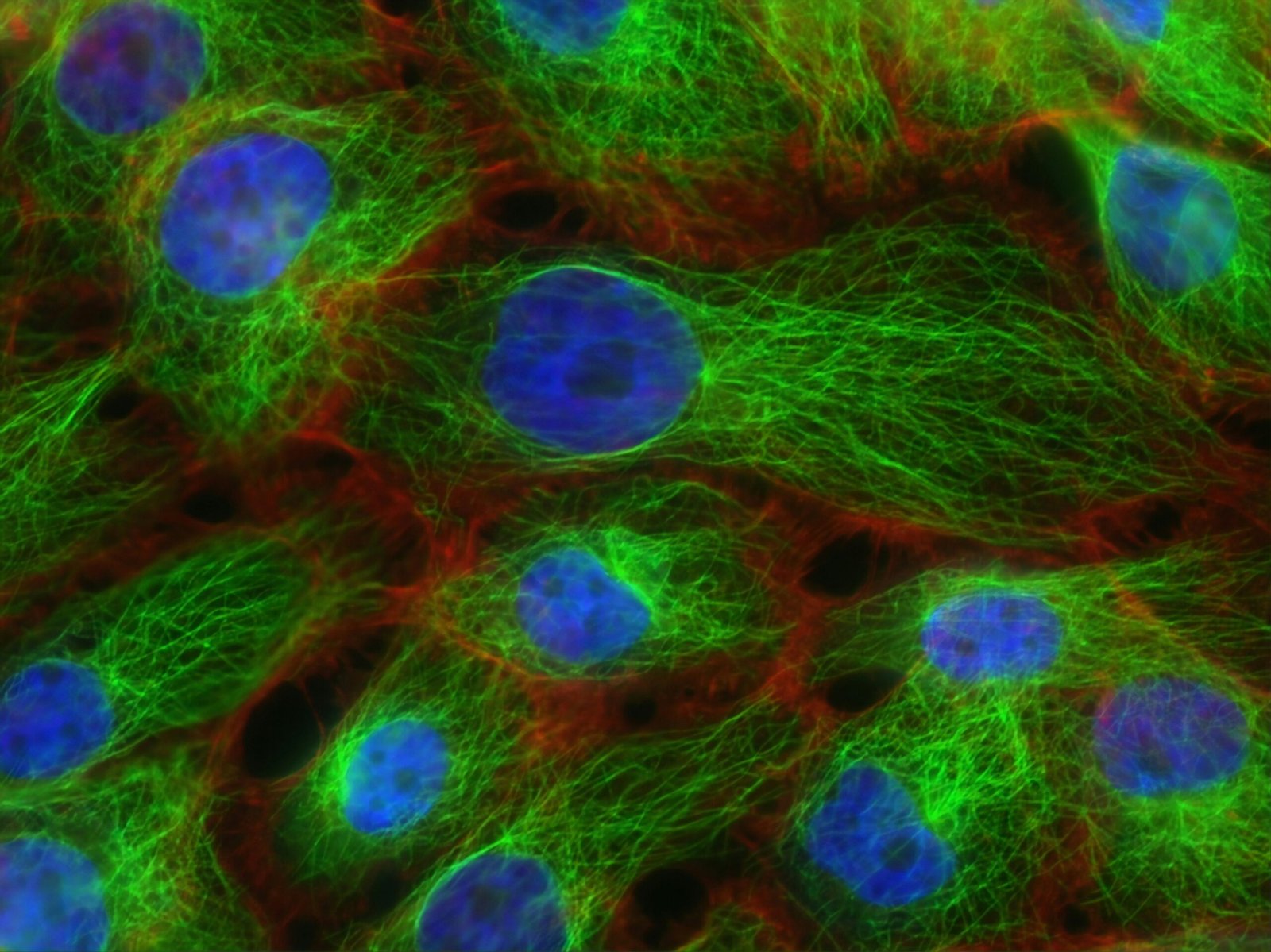
Genome-wide analyses of gene families in plants rely on a suite of bioinformatics methodologies that facilitate the identification and classification of these gene families. Essential to these analyses are sequence alignment techniques, which enable researchers to compare gene sequences across various species. By employing tools such as BLAST (Basic Local Alignment Search Tool), researchers can identify homologous sequences, revealing evolutionary relationships among genes. These sequences can then be aligned using programs like Clustal Omega, allowing for the detection of conserved regions that are crucial for understanding gene family dynamics.
Phylogenetic analysis serves as another critical methodology in the study of gene families. It utilizes the aligned sequences to construct phylogenetic trees, which visualize the evolutionary connections among different genes. Tools like MEGA (Molecular Evolutionary Genetics Analysis) facilitate this process by providing algorithms for both distance-based and character-based methods, helping to elucidate the evolutionary history and functional divergence of gene families across the plant kingdom.
Gene annotation is another vital aspect of genome-wide analyses. This process involves assigning functional information to genes, which can be achieved through high-throughput sequencing technologies, such as RNA-Seq. These technologies generate vast amounts of genomic data that are indispensable for understanding the expression patterns of different gene families. Additionally, databases like GenBank and Ensembl Plants serve as repositories for storing this genomic data, providing a resource for researchers to access and share information.
Practical examples of these methodologies can be observed in studies of specific plant species. For instance, genome-wide analyses of the Arabidopsis thaliana genome have employed these methodologies extensively, leading to significant insights into its gene families. Similarly, the application of these bioinformatics tools has advanced our understanding of crops like rice and maize, highlighting the importance of these techniques in advancing plant genomics.
Case Studies: Insights from Recent Research
Recent studies into plant gene families have yielded significant insights, revealing the intricate roles these genetic units play in various plant species. One notable case study examined the gene families associated with drought resistance in Arabidopsis thaliana. Through genome-wide analysis, researchers identified a specific gene family, termed the COR (Cold-Regulated) gene family, which plays a crucial role in how the plant responds to water scarcity. The research demonstrated that members of this gene family were upregulated under drought conditions, effectively allowing the plant to conserve water and maintain cellular functions. These findings underscore the potential of manipulating the COR gene family to enhance drought resilience in economically important crops, thereby aiding agricultural sustainability.
Another significant case study focused on the major gene families linked to disease resistance in Oryza sativa (rice). The work highlighted the role of the NBS-LRR (Nucleotide-binding site-Leucine Rich Repeat) gene family, which is pivotal in plant defense mechanisms against biotic stressors such as pathogens. The comprehensive genome-wide analysis reported a rapid evolution of this gene family, highlighting its adaptability and functional diversity. By employing marker-assisted selection strategies based on NBS-LRR gene variants, plant breeders can potentially create rice varieties with enhanced resistance to prevalent diseases, thus improving food security in regions where rice is a staple food.
Further research into the Met gene family in coconuts revealed not only their importance in metabolic regulation but also their influence on traits relevant to plant fitness, such as growth patterns and yield. The study elucidated how variations within this gene family could lead to substantial differences in phenotypic outcomes, which holds promise for agricultural practices aiming to boost yield and adaptability in changing climates.
These case studies illustrate the profound implications of genome-wide analyses of gene families for advancing our understanding of plant biology, agricultural practices, and plant breeding strategies. The connection between genetic insights and practical applications presents a pathway for significant advancements in plant sciences and their related fields.
Future Directions and Challenges in Gene Family Research
As the field of plant genomics continues to evolve, the future of gene family research holds considerable promise and presents various challenges. One emerging trend is the integration of genomics with phenomics and transcriptomics. By combining these different approaches, researchers can achieve a more comprehensive understanding of gene function and regulation. This multi-faceted approach allows for the investigation of how specific gene families interact with environmental factors, thereby influencing phenotypic traits. As technology advances, high-throughput sequencing combined with detailed phenotypic assessments will likely enhance the resolution of gene family analyses and their associated functions.
Despite these promising directions, several challenges persist in the domain of gene family research. One major challenge is the complexity involved in gene family evolution. Gene duplication events, rapid diversification, and loss of gene function complicate the reconstruction of gene family phylogenies. Understanding the evolutionary trajectories of these gene families requires sophisticated analytical techniques and robust computational tools capable of handling vast datasets. Additionally, functional validation of gene family members remains difficult, as many genes exhibit redundant functions within their families. This redundancy can obscure the specific roles of individual genes in biological processes.
Furthermore, the need for more diverse plant genomic resources cannot be overstated. Many existing databases mainly focus on model organisms, which may not accurately represent the genetic diversity found in non-model plant species. Expanding genomic resources to include a broader range of species will enhance our understanding of gene family dynamics across different plant lineages. This expansion is crucial for translating genomic insights into practical applications, particularly in crop improvement and sustainable agriculture.
Advances in gene family research have the potential to unlock new strategies for enhancing crop resilience and sustainability. By elucidating the complex roles of gene family members, researchers can develop more targeted breeding programs and biotechnological interventions aimed at improving yield and stress tolerance. Thus, addressing the challenges and harnessing the emerging trends in this field will be essential for future advancements in agricultural science.






Однажды в сказке
На этом сайте можно найти информацией о телешоу "Однажды в сказке", развитии событий и ключевых персонажах. https://odnazhdy-v-skazke-online.ru/ Здесь размещены подробные материалы о создании шоу, исполнителях ролей и фактах из-за кулис.
Denniswor
Программа наблюдения за объектами – это актуальное решение для защиты имущества, объединяющий технологии и удобство использования . На веб-ресурсе вы найдете детальные инструкции по настройке и установке систем видеонаблюдения, включая онлайн-хранилища, их сильные и слабые стороны. Программа видеонаблюдения Рассматриваются комбинированные системы, сочетающие облачное и локальное хранилище , что делает систему универсальной и эффективной. Важной частью является разбор ключевых интеллектуальных возможностей, таких как определение активности, распознавание объектов и дополнительные алгоритмы искусственного интеллекта.
Exploring The Final Syllabus Of Plant Molecular Techniques
[…] Exploring Gene Families in Plants: A Genome-Wide Analysis […]
Video Surveillance Software
This website provides a valuable overview of various desktop Video Surveillance Software options, and the emphasis on free IP camera video surveillance solutions is greatly appreciated. The inclusion of AI-powered object detection capable of differentiating between people, cats, birds, and dogs represents a significant advancement in security technology, especially as they say that the functionalities are accessible without cost. I look forward to reading user reviews and gaining insights into the overall performance and usability of these software packages, which are available for IP camera recording and come with time-lapse recording.