What are Protein Domains?
Within a protein, protein domains are discrete structural and functional components. They can be modular, which means they can show up in different proteins, and each domain can support a certain function. Researchers can determine the protein’s function and comprehend the evolutionary processes that resulted in its current condition by examining these domains. Protein domain recognition can help predict the function of uncharacterized genes based on the domains they include in genome-wide investigations.
The Role of Protein Motifs
Protein domains are made up of motifs which are very brief sequences that are shared by all proteins and frequently associated with certain functions. For example, a basic motif might be essential for binding or signaling. Since these patterns reveal a protein’s function throughout biological pathways, their identification is crucial for genome-wide study. Our comprehension of intricate biological processes can be improved by this knowledge.
Utilizing Genome-Wide Analysis
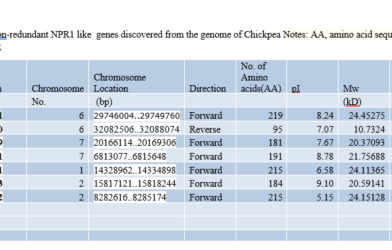
By thoroughly examining an organism’s whole genetic makeup, genome-wide analysis allows researchers to link particular protein domains and patterns to particular gene activities. Researchers can identify patterns that show how genes contribute to disease or other biological processes by analyzing large volumes of data using computational techniques and high-throughput sequencing. This comprehensive approach is crucial to improving our understanding of gene activity and how it affects health and illness.
For Power point prsentation on understanding the motif and domain in protein please click here
Understanding Motif Finder for Protein Domain Identification in Genome-Wide Analysis







What Is Motif And Domain In Protein -
[…] Protein Domains and Motifs prediction in Genome-Wide Analysis […]